Colossal-AI and Intel® Partner to Deliver Cost-Efficient Open-Source Solution for Protein Folding Structure Prediction with Habana Gaudi Processors

The Colossal-AI team and Intel’s Habana® Labs collaborated to optimize the performance of protein structure prediction when using DeepMind’s AlphaFold2 model. Colossal-AI improved the end-to-end inference speed by up to 3.86 times compared to existing solutions, and with Habana Gaudi processors, the application cost was reduced by up to 39% compared to GPU.You can find the open-source code at https://github.com/hpcaitech/ColossalAI#biomedicine
DeepMind's AlphaFold2 artificial intelligence technology can accurately predict protein structures based on amino acid sequences. It was selected as one of the top ten scientific breakthroughs in 2021 by Science and Nature. Since then, the field of life sciences has seen a surge in the use of artificial intelligence for protein structure prediction, which has been widely applied in biological research and drug development. However, the high economic and time costs of using AlphaFold2 have posed serious obstacles to related research and applications.
Colossal-AI is the most popular open-source large AI model solution repository with nearly 20,000 GitHub Stars. The repository has showcased outstanding performance in a variety of large models, including ChatGPT, LLaMA, Stable Diffusion, GPT-3, OPT, and PaLM. In the case of AlphaFold2, Colossal-AI has utilized advanced technologies such as dynamic axial parallelism, kernel fusion, asynchronous distributed pre-processing, and block computing to significantly accelerate training and inference time, reduce GPU memory usage, and lower application costs.
Colossal-AI and Intel Habana have joined forces to offer a new AI solution for the life science industry. This joint solution leverages the Habana® Gaudi® processor, which is specifically designed for high-performance and high-efficiency Deep Learning (DL) training and inference, while providing developers with optimized software and tools that can scale to handle various workloads and systems.
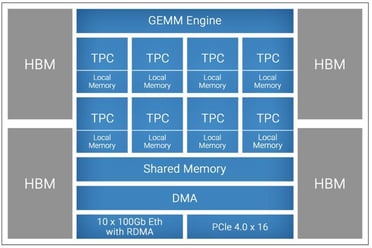
Figure 1. First-gen Gaudi processor high-level architecture
The Gaudi processor architecture is composed of three main subsystems - compute, memory, and networking. The compute architecture is heterogeneous and includes two compute engines – a GEMM engine, also called Matrix Multiplication Engine (MME), and a fully programmable Tensor Processor Core (TPC) cluster. The MME is responsible for performing all operations that can be reduced to Matrix Multiplication (such as fully connected layers, convolutions, batched-GEMM), while the TPC, a VLIW SIMD processor specifically designed for deep learning operations, is used to accelerate everything else. Additionally, the Gaudi architecture is also the first DL training processor that has integrated ten RDMA over Converged Ethernet (RoCE v2) engines on-chip. Based on the Gaudi processor architecture, Habana has two generations of products being sold in the market. These two generations of products follow the same architecture and software, providing customers with the same user experience but different performance.
Colossal-AI and Intel Habana have collaborated to optimize the AlphaFold2 model for protein folding structure prediction. The team leveraged Colossal-AI for large AI model optimization while using Intel’s 3rd Gen Intel® Scalable Xeon® CPUs for data preprocessing and performed model inference on Habana Gaudi processors.
Performance Data Comparison
Data preprocessing on Xeon processors with Colossal-AI
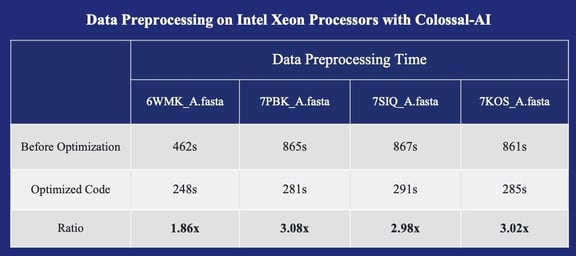
Hardware Configuration: two Intel(R) Xeon(R) Platinum 8368 CPU @ 2.40GHz, and 1TB of System Memory
Inference on Intel Habana Gaudi with Colossal-AI
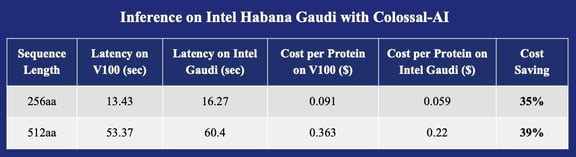
* Cost based on AWS on-demand pricing for P3.16xlarge instance and Gaudi-based DL1.24xlarge instance. https://aws.amazon.com/en/ec2/pricing/on-demand/
Colossal-AI is partnering with Intel’s Habana Labs to optimize inference on Gaudi2 processors. The partnering teams are also working to enable training on both first-generation Gaudi and Gaudi2 processors.
Model Migration Best Practices
The process of migrating Colossal-AI to Intel Habana Gaudi follows the official documentation, using the Docker environment equipped with the SynapseAI® Software Suite 1.7.1 environment as the development and test environment. The overall migration process can be divided into the following steps:
1. Porting the model to the Gaudi platform is simple and easy.
a. Import the Intel Habana Torch library: import habana_frameworks.torch.core as htcore
b. Specify the device as hpu: device = torch.device("hpu")
c. Add htcore.mark_step(), to trigger graph compilation and execution
d. Use reshape+matmul to re-implement the einsum of some high-dimensional tensors.
2. Enabling distributed inference and training is simplified with MPI and Intel Habana’s HCCL library.
a. Use MPI to start multiple processes, and use mpi4py to obtain distributed information (world_size, rank)
b. Specify HCCL (Habana Collective Communication Library) as the communication backend.
# Python
import habana_frameworks.torch.distributed.hccl
dist.init_process_group(backend='hccl', rank=rank, world_size=world_size)
3. The Habana HMP (Habana Mixed Precision) package enables mixed precision (bfloat16 and float32) during training and inference.
a. Before the training loop or inference starts, add the following lines to enable mixed precision
# Python
from habana_frameworks.torch.hpex import hmp
hmp.convert('O1') b. Use hmp.disable_casts to disable mixed precision in part of the code (FeaturePipeline, Optimizer)
When debugging code, using eager mode can help locate errors more conveniently and provide additional information for debugging. To run the code in eager mode, set the environment variable PT_HPU_LAZY_MODE to 2. Some environment variables related to LOG can also be set to output more debugging information:
# Bash
export LOG_LEVEL_ALL=4
export ENABLE_CONSOLE=trueOn the Gaudi platform, the compilation process is triggered each time the model is run, which will reduce the efficiency of development or debugging. To avoid repeated compilation and speed up the development or debugging process, the path of compilation to the result cache can be specified through the environment variable PT_RECIPE_CACHE_PATH.
Performance optimization
Colossal-AI has integrated its large model optimization technology into AlphaFold2's training and inference, which reduces the economic and time costs. These improvements leverage the excellent performance and efficiency of Habana Gaudi processors and the new 3rd generation Intel® Xeon® processors as follows:
1. Colossal-AI utilizes computing optimization technologies such as operator fusion and re-implements LayerNorm and Fused Softmax according to performance characteristics of the AlphaFold2 model, thus greatly improving its computing efficiency on acceleration devices. To analyze the performance of the model and find the main performance bottlenecks, rich and free performance optimization software stack and tools were used which are available with the Habana Gaudi platform, as shown in Figure 2:
Figure 2. Profile before optimization
By utilizing the custom op tool and method provided by Habana to implement the Fused Softmax operator running on the TPC, the partnering teams were able to improve the execution efficiency of the operator and the increase the utilization rate of the computing engine, as depicted in Figure 3:
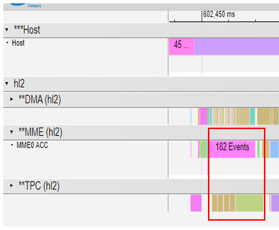
Figure 3. Profile after Optimization
2. Colossal-AI utilizes Ray as a distributed computing engine to support asynchronous distributed calls. For protein structure inference of a single amino acid sequence, the partnering teams have achieved approximately 3 times the pre-processing acceleration. For protein complex structure inference composed of N amino acid sequences, they have achieved about 3*N times the pre-processing acceleration, which significantly reduces the cost of deploying AlphaFold. The preprocessing code mainly consumes resources such as CPU and disk capacity, and the optimization approach is as follows:
(1) Optimize two key MSA (Multiple Sequence Alignment) loads based on the Intel intrinsics instruction set: jackhmmer and hhblits. Use Intel® VTune to analyze the hotspots, introduce AVX512 high-bit-width SIMD functions and MSA multi-process concurrency, and compile with Intel®. Compile the source code with icc (enable option -O3 -march=icelake-server).
(2) Use numactl -m <memory NUMA number> -C <physical core number list> to bind threads, execute multiple instances in parallel, and let the CPU utilization rate reach 100%, in order to obtain maximum throughput optimization.
3. In addition, Colossal-AI supports model parallelism and distributed inference during training through DAP (Dynamic Axial Parallelism). During actual deployment, DAP can distribute computation to multiple devices, significantly reducing training and long-sequence inference time. Colossal-AI proposes to furthermore reconstruct and optimize the block computing technology, adjust each module in a targeted manner, optimize the logic and efficiency of a block, and reduce the peak memory usage of the computing module by 40% during long sequence inference.
Eitan Medina, Chief Operating Officer of Habana Labs, an Intel company, said, “The Habana team is pleased to collaborate with Colossal-AI and Intel to deliver AlphaFold2, which brings significantly faster and lower cost inference to protein folding research, making it more accessible to a broader array of research initiatives. “
Professor Yang You, Chairman and President of HPC-AI Tech, stated, "The jointly developed AlphaFold2 solution by Colossal-AI and Intel Habana is an important milestone for the future of biomedicine. Colossal-AI and Intel are the leaders in AI infrastructure in the field of biomedicine. In the future, the two parties will continue to deepen their cooperation in frontier fields such as AIGC and AI for Science, helping to reduce costs and increase the efficiency of large AI model applications."
You can find the open-source code at https://github.com/hpcaitech/ColossalAI#biomedicine
Appendix
About Colossal-AI
Colossal-AI is a deep learning system for the large AI model era, which supports efficient and fast training and inference of large AI models based on PyTorch, and reduces the cost of large AI model deployment. Since becoming open-source, Colossal-AI has ranked first on the GitHub Trending multiple times and has gained almost 20,000 GitHub Stars. It has been chosen as the official tutorial for top AI and HPC conferences such as SC, AAAI, PPoPP, CVPR and ISC. Its solutions have been successfully applied and acclaimed by well-known tech giants in the fields of autonomous driving, cloud computing, retail, medicine, chips, etc. For example, Colossal-AI has successfully helped a Fortune-500 enterprise develop a ChatGPT-like chatbot model with enhanced online searching capabilities.
About Habana
Habana Labs, an Intel company, delivers purpose-built processor platforms optimized for high-performance, high-efficiency deep learning training and inference of large-scale models. Habana provides solutions for cloud service providers as well as enterprise customer on-premises deployments.
About Intel Habana Gaudi2 AI Processors
A new generation of Habana Gaudi2 processors raises deep learning training and inference performance to a whole new level and is available now for cloud and on-premises deployment. In terms of specific workload performance, taking ResNet-50 and BERT as examples, thee models achieve twice the throughput of A100, as shown in Figure 4.
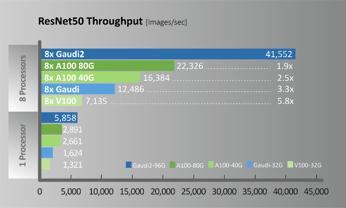
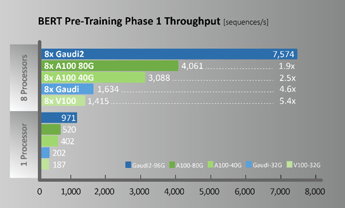
Figure 4. ResNet50 / BERT Performance comparison (Configuration for throughput measurements provided below)
For more specific product information, please visit https://habana.ai, or visit the Intel Developer Cloud experience. For more developer information, please visit https://developer.habana.ai
Citations
- (6aa)PDBDOI: http://doi.org/10.2210/pdb6WMK/pdb Chain A of beta sheet heterodimer LHD29
- (646aa)PDDOI: http://doi.org/10.2210/pdb7PBK/pdb Chain A of Vibriophage phiVC8 family A DNA polymerase (DpoZ)
- (542aa)PDBDOI: http://doi.org/10.2210/pdb7SIQ/pdb Chain A of a peptide chain release factor 3 (prfC) from Stenotrophomonas maltophilia
- (149aa)PDBDOI: http://doi.org/10.2210/pdb7KOS/pdb Chain A of Putative Pterin Binding Protein PruR (Atu3496)
- Colossal-AI open-source address: https://github.com/hpcaitech/ColossalAI
- Habana SynapseAI Container: https://vault.habana.ai/ui/repos/tree/General/gaudi-docker/1.7.0/ubuntu20.04/habanalabs/tensorflow-installer-tf-cpu-2.8.3
- Habana Gaudi Performance: https://developer.habana.ai/resources/habana-training-models/
- https://github.com/deepmind/alphafold
- Configuration data for Gaudi2 comparative measurements:
Habana ResNet50 Model: https://github.com/HabanaAI/Model-References/tree/master/TensorFlow/computer_vision/Resnets/resnet_keras
Habana SynapseAI Container: https://vault.habana.ai/ui/repos/tree/General/gaudi-docker/1.7.0/ubuntu20.04/habanalabs/tensorflow-installer-tf-cpu-2.8.3
Habana Gaudi Performance: https://developer.habana.ai/resources/habana-training-models/
A100 / V100 Performance Source: https://ngc.nvidia.com/catalog/resources/nvidia:resnet_50_v1_5_for_tensorflow/performance, results published for DGX A100-40GB and DGX V100-32GB
Measured January 2023.
Habana BERT-Large Model: https://github.com/HabanaAI/Model-References/tree/master/TensorFlow/nlp/bert
Habana SynapseAI Container: https://vault.habana.ai/ui/repos/tree/General/gaudi-docker/1.7.0/ubuntu20.04/habanalabs/tensorflow-installer-tf-cpu-2.8.3
Habana Gaudi Performance: https://developer.habana.ai/resources/habana-training-models/
A100 / V100 Performance Sources: https://ngc.nvidia.com/catalog/resources/nvidia:bert_for_tensorflow/performance,
results published for DGX A100-40G and DGX V100-32G Measured January 2023
Results may vary.
Comments